Nuclic Acids and Enzymes
Table of Content |
Nucleic Acids
 Meischer discovered nucleic acids in nucleus of pus cell and called it nuclein. The name nucleic acid proposed by Altman.
Meischer discovered nucleic acids in nucleus of pus cell and called it nuclein. The name nucleic acid proposed by Altman.
Nucleic acids are polymer nucleotides. = Nitrogen base + pentose + phosphate
On the, basis of structure nitrogen bases are broadly of two types:
Pyrimidines: Consist of one pyrimidine ring.
Skeleton of ring composed of two nitrogen and four carbon atoms.
e.g. Cytosine, Thymine and Uracil.
Purines: Consist of two rings i.e. one pyrimidine, ring (2N + 4C) and one imidazole ring (2N + 3C) e.g adenine and guanine.
Pentose Sugar:
Nitrogen base forms bond with first carbon of pentose sugar to form a nucleoside, nitrogen of third place (N3) forms bond with sugar in case of pyrimidines while in purines nitrogen of ninth place (N9) forms bond with sugar.
Phosphate forms ester bond (covalent bond with fifth carbon of sugar to form a complete nucleotide.
Types of Nucleosides and Nucleotides
Adenine + Ribose = Adenosine
Adenosine + Phosphate = Adenylic acid
Adenine + Deoxyribose = Deoxy adenosine
Deoxy adenosine + P = Deoxy adenylic acid
Guanine + Ribose = Guanosine
Guanosine + P = Guanylic acid
Guanine + Deoxyribose = Deoxy guanosine
Deoxy guanosine + P = Deoxy guanylic acid
Cytosine + Ribose = Cytidine
Cytidine + P = Cytidylic acid
Cytosine + Deoxyribose = Deoxycytidine
Deoxycytidine + P = Deoxycytidylic acid
Uracil + Ribose = Uridine
Uridine + P = Uridylic acid
Thymine + Deoxyribose = Deoxy thymidine
Deoxythymidine + P = Deoxythymidylic acid
DNA
Discovered by - Meischer
DNA term given by - Zacharis
In DNA pentose sugar is deoxyribose sugar and four types of nitrogen bases A, T, G, C.
Wilkins and Franklin studied DNA molecule with the help of X-Ray crystallography.
With the help of this study, Watson and Crick (1953) proposed a double-helix model for DNA. For this model Watson, Crick and Wilkins were awarded noble prize in 1962.
According to this model, DNA is composed of two polynucleotide chains.
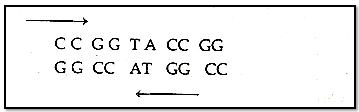
Both polynucleotide chains are complementary and anti parallel to each other.
In both strand of DNA direction-of phosphodiester bond is opposite, i.e. if direction of phosphodiester bond in one strand is 3'-5" then it is 5'-3'" in another strand.
Both stran4 of DNA held together by hydrogen bonds. These hydrogen bonds are present between nitrogen bases of both strand.
Adenine binds to Thymine by two hydrogen bonds and Cytosine binds to Guanine by three hydrogen bonds.
Chargaff's equivalency rule: In a double stranded DNA amount of purine nucleotides is equals to amount of pyrimidine nucleotides.
Purine = Pyrimidine
[A] + [G] = [T] + [C]
Base ratio
In a DNA, A + T > G + C Þ A - T type DNA.
Base ratio of A - T type of D~A is more than one. e.g. Eukaryotic DNA
In a DNA, G + C > A + T Þ G - C type DNA.
Base ratio of G - C type of DNA is less than one. eg. Prokaryotic DNA
Melting point of DNA depends on G - C contents.
More G - C contents then more melting point.
Tm of prokaryotic DNA > Tm of Eukaryotic DNA
DNA absorbs U.V. rays means 2600A wavelength.
Out of two strand of DNA only one strand participates in transcription, it is called antisense strand/non coding strand/template strand.
Other strand of DNA which does not participate in transcription is called sense strand/ coding strand.
Configuration of DNA Molecule
Two strands of DNA are helically coiled like a revolying ladder. Back bone of this ladder (Reiling) is - composed of phosphates and sugars while steps (bars) composed of pairs of nitrogen bases.
Distance between two successive steps is 3.4 A, In one complete turn of DNA molecule there are such 10 steps (10 pairs of nitrogen bases). So the length of one complete turn is 34 Å. This is called helix length.
Diameter of DNA molecule i.e distance between phosphates of two strands is 20Å.
Distance between sugar of two strands is 11.1Å.
Length of hydrogen bonds between nitrogen bases is
2'.8-3.0Å. Angle between nitrogen base and C1 Carbon of pentose is 51°.
Molecular weight of DNA is 106 to 109 dalton.
In nucleus of eukaryotes the DNA is associated with 'histone protein to form nucleoprotein. Histone occupies major groove of DNA at 30° angle.
Bond between DNA and histone is salt linkage (Mg+2).
DNA in chromosomes is linear while in prokaryotes, mitochondria and chloroplast it is circular.
In f × 174 bacteriophage the DNA is single stranded and circular isolated by Sinsheimer.
G-4, S-13, M-13, F1 and Fd - Bacteriophages also contain ss-circular DNA.
Types of DNA
On the basis of direction of twisting, there are two types of DNA.
Right Handed DNA
Clockwise twisting e,g. The DNA for which Watsor and Crick proposed model was B DNA.
Other e.g. of right handed DNA
|
DNA |
Helix Length |
No. of base pairs |
Distance b/w two |
Pairs Diameter |
|
A |
28 Å |
11 pairs |
2.56 Å |
23 Å |
|
B |
34 Å |
10 pairs |
3.4 Å |
20 Å |
|
C |
31 Å |
9.33pairs |
3.32 Å |
19 Å |
|
D |
24.2 Å |
8 pairs |
3.03 Å |
19 Å |
Left Handed DNA:
Anticlockwise twisting e.g. Z-DNA-discovered by Rich.
Phosphate and sugar backbone is zig-zag.
Units of Z-DNA are dinucleotides (purine and pyrimidine in alternate order)
Helix length 45.6 Å, Diameter - 18.4 Å
No. of base pairs 12 (6 dimers)
Distance between base - pairs. = 3.75 Å
Palindromic DNA: - Wilson and Thomas
Sequence of nucleotides same from both ends.
NOTE: DNA molecule is Dextrorotatory while RNA molecule is Laeyorotatory.
C-value = Total amount of DNA in a haploid genome of organism.
RIBO Nucleic Acid (RNA)
Structure of RNA is fundamentally the same as DNA, but there are some differences. The differences are:
In place De -oxyribose sugar of DNA, there is present Ribose sugar in RNA.
In place of nitrogen base Thymine present in DNA, there is present nitrogen base uracil in-RNA.
RNA is made up of only one polynucleotide chain i.e. R.N.A. is single stranded.
Exception: RNA found in Reo-virus is double stranded. i.e. it has two polynucleotide chains.
Types of RNA
1. Genetic RNA or Genomic RNA:
In the absence of DNA, sometime RNA working as genetic material and genomic RNA transfer in formations from one generation to next generation.
E.g. Reo virus, TMV, QB bacteriophage,
2. Non-genetic RNA - 3 types
(1) r - RNA
(2) t - RNA
(3) m - RNA
(1) Ribosomal RNA (r - RNA)
This RNA is 80% of the cells total RNA
It is found in ribosomes and it is produced in nucleolus,
It is the most stable form of RNA.
There are present 80S type of ribosomes in Eukaryotic cell. Their subunits are 60S and 40S. In 60S sub unit of ribosome three types of r-RNA are found 5S, 5.8S, 28S.
In the same way 40s sub unit of ribosome has only one type of r-RNA = 18S.
80S ribosome has total 4 molecules of r-RNA.
Prokaryotic cells have 70S type of ribosomes and its subunits are 50S and 39S.
50S sub unit of ribosome contains 2 molecules of rRNA = 5S and 23S.
30S sub unit of ribosome has 16S type of r-RNA.
70S RNA has total 3 molecules of r-RNA.
Function: At the time of protein synthesis r-RNA provides attachment site 'to t-RNA and m-RNA and attaches them on the ribosome.
The bonds formed between them are known as salt linkages. It attached – RNA to the larger subunit of ribosome and m-RNA to smaller subunit of ribosome.
(2) Transfer: - RNA (t-RNA)
It is 10-15% of total RNA.
It is, synthesized in the nucleus by DNA.
It is also known as soluble RNA (sRNA)
Function: At the time of protein synthesis it acts as a carrier of amino-acids.
Discovery: t-RNA was discovered by Hogland, Zemecknike and Stephenson.
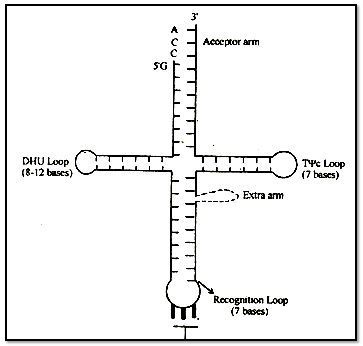
Structure: The structure of t-RNA is most complicated.
A scientist named Holley presented Clover leaf mood of its structure. In two dimensional structure the t-RNA appears cover leaf like but in three dimensional structure (by Kim) it appears L-shaped.
The molecule of t - RNA is of single strand.
There are-present three nucleotides in a particular sequence at 3' end- of t - RNA and that sequence is = CCA.
All the 5. ends i.e. last ends-are having G (guanine).
3' end is known as Acceptor end.
t-RNA accepts amino acids -at acceptor points.
Amino acid-hinds to 3' end by its - COOH group.
The molecule of t-RNA is folded and due to folding some-complementary nitrogenous bases come across with each other and form hydrogen bonds.
There are some places where hydrogen bonds are not formed, these places are known as loop. Loops: There are some abnormal nitrogenous bases in the loops, that is why hydrogen bonds are not formed.
e.g. Inosine (I) Pseudouracil (ψ), Dihydrouridine (DHU)
(A) T ψ C Loop or Attachment loop :
This loop connects t - RNA to the larger subunit of ribosome.
(B) Recognition Loop:
This is the most specific loop of t - RNA and different types of t - RNA are different due to this loop. There is a specific sequence of three nucleotides called anticodon, is present at the end of this loop.
On the basis of anticodon, there are total 61 types of t- RNA or, we can also say that there are 61 types of anticodon.
t-RNA recognizes its place on m-RNA with the help of anticodon.
The anticodon of t-RNA recognises its complimentary sequence on m-RNA. This complimentary sequence is known as codon.
(C) DHU Loop:
It is also known as Amino - acyl synthetase recognition loop. Amino-acyl synthetase is a specific type of enzyme. The function of this enzyme is to activate a specific type of amino acid after activation this enzyme attaches the amino acid to the 3' end of t-RNA.
There are 20 types of enzymes for 20 types of aminoacids.
The function of DHU loop is to recognize this specific Aminoacyl synthetase enzyme.
(3) Messenger RNA (m -RNA):
The m - RNA is 1-5% of the cell's total RNA
Discovery: Messenger RNA was discovered by Huxley, Volkin and Astrachan. The name m-RNA was given by Jacob and Monad.
The m - RNA is produced by genetic DNA in the nucleus. This process is known as Transcription
Enzyme
Term enzyme was given by Kuhne.
Zymase (from yeast) was the first discovered enzyme by Buchner.
Enzymes are biocatalysts made up of proteins (except ribozyme), which increases the rate of biochemical reactions by lowering down the activation energy.
The first purified and crystalized enzyme was urease (by J.B.Sumner) from Canavalia/Tack Bean (Lobia plant).
Proteinaceous nature of enzyme was suggested by Northrop and Sumner.
First discovered ribozyme was L19 RNA ase by T.Cech from rRNA of a protozoan Tetrahymena thermophila and RNAase P or Ribonuclease P by Altman in prokaryotic cell,
Characteristics of Enzymes
All enzymes are proteins, but all proteins are no enzymes.
All enzymes are tertiary & globular-proteins (Isoenzymes quarternary protein)
Enzymes accelerate the rate of reaction, without undergoing any change in themselves. Enzymes lower down the activation energy of substrate or reactions.
Enzymes are macromolecules of amino acids, which are synthesized on ribosomes under the control of genes.
M.wt. of enzymes are high and they are colloidal substances.
Enzymes are very sensitive to pH & temperature.
Optimum temp for enzymes is 20-35°C.
Most of enzymes are active at neutral pH, hydrolytic enzymes of lysosomes are active on acidic pH (5).
Enzymes are required in very minute amount for bio-chemical reactions.
Their catalytic power is represented by Michaelis
Menten constant or Km constant and turn over number.
"The number of substrate molecules converted into products per unit time by one molecule of the enzyme in favourable conditions is called turn over number." The maximum turn over number is of Carbonic anhydrase is 360 lakh, for Catalase is 50 lakh, for favoprotein is 50 & for lysozyme is 30 per minute.
Enzymes are very specific to their substrate or reactions.
Structure of Enzyme
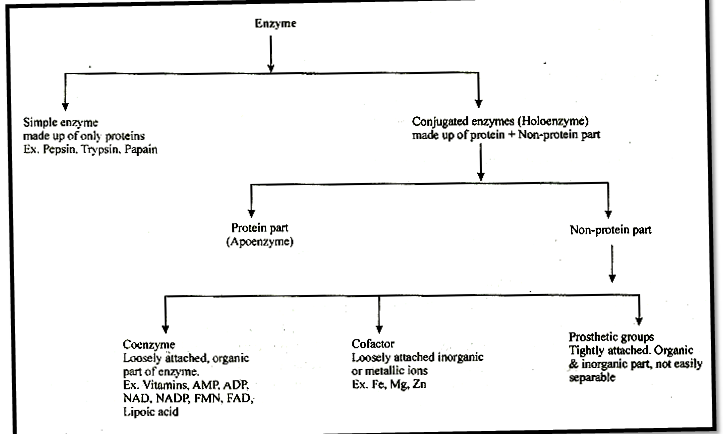
Simple enzyntes: They are made up of only proteins e.g pepsin, trypsin, papain.
Conjugated enzymes: They are made up of a protein part & non protein part.
(i) Co-enzymes: Co-enzymes are non-protein organic groups, which are loosely attached to apoenzymes. They are generally made up of vitamins.
(ii) Prosthetic group: When non-protein part is tightly or firmly attached to apoenzyme.
(iii) Metal activators/co-factors metallic factor:
Loosely attached inorganic co-factor. E.g. Mn, Fe, Co, Zn, Ca, Mg, Cu.
Active site: Specific part of amino acid chain in enzyme structure at which specific substrate is to be binded and catalysed is known as active site. Active site of enzyme is made up of very specific sequence of amino acids, determined by genetic codes.
Allosteric site: Besides the active site's, some enzymes possess additional sites, at which chemicals other than substrate (allosteric modulators) are bind. These sites are known as allosteric sites and enzyme with allosteric sites are called as allosteric enzymes. e.g. hexokinase, phosphofructokinase.
Terminology
Endoenzymes: Enzymes which are functional only inside the cells.
Exoenzymes: Enzymes catalysed the reactions outside the cell Eg:- enzymes of digestion, some enzymes of insectivorous plants, Zymase complex of fermentation.
Proenzyme/Zymogen: These are precursors of enzymes or inactive forms of enzymes.
E.g. Pepsinogen, Trypsinogen etc.
Iso-enzymes: Enzymes having similar action, but little difference in their molecular configuration an called isoenzymes. 16 forms of a-amylase of wheat & 5 forms of LDH (Lactate dehydrogenase) art known. These all forms are synthesised by different genes.
Inducible enzymes: When formation of enzyme is induced by substrate availability.
e.g Lactase, Nitrogenase, b-galactosidase.
Extremozymes: Enzymes, which may also function at extremely adverse conditions e.g Tae polymerase.
Abzymes: When the monoclonal antibodies an used as enzymes or reagents.
Biodetergents: Enzymes used in washing powders are known as bio-detergents e.g. amylase, lipase, proteolytic enzymes.
House keeping / constitutive enzymes: Which are always present in constant amount & are also essential to cell.
Nomenclature and Classification
Enzyme commission of IUB-1961 divides all enzymes into 6 major classes and also proposed an international code of 4 digits for each enzyme,
(I) Oxido-reductases: These enzymes involve in oxidation-reduction reactions. It involves 3 sub classes (i) Oxidase (ii) dehydrogenase (iii) reductase
E.g.- Cytochrome oxidase.
(II) Transferases: These enzymes transfer specific group from one substrate to another.
E.g.- Transaminase, Hexokinase.
(III) Hydrolases: These Enzymes involve in hydrolysis reactions with help of H2O.
E.g. Proteases,' Lipases, Carbohydrases,
(IV) Lyases: Split the substrate molecule without water These Enzymes splits the specific covalent bond:
Without hydrolysis or H20 addition. E.g. Aldolase
(V) Isomerases: Rearrangement of molecular structure to form isomers.
(VI) Ugases (Synthases): Covalent bonding of two substrates to form a large molecule. E.g. Citrate synthetase, Ligase
Mode of Action of Enzyme
1. Lock & Key theory or template theory:
Given by Emil Fischer
According to this theory active sites of enzymes serve as a lock into, which the reactant substrate fits like a key.
Supported by competitive inhibition.
2. Enzyme - substrate complex theory:
Given by Henry explained by Michaelis & Menten Enzyme + Substrate ® ESC ® EPC ® Product + Enzyme
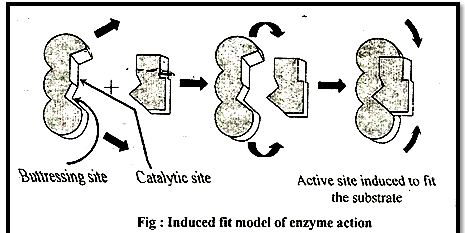
3. Induced fit theory:
Given by D.E. Koshland (1973-74)
According to this theory active site is not static-, but it undergoes a conformational change which is induced by specific substrate. The active site has two groups, (a) Buttressing (supporting) group & (b) Catalytic group - Buttressing group is meant for supporting the substrate, while the catalytic group break the substrate into product.
Factors
pH: Enzymes very sensitive to pH.
Temperature: High temp inactivates enzyme causing their denaturation. They also get inactive at lower temp. Generally all enzymes better perform at body temp of organism.
Enzyme concentration: Increase in cone. of enzymes will increase the rate of enzymatic reaction till enough the substrate,
Substrate concentration: Increase in substrate cone" increases the activity of enzymes until all the active sites of enzyme are saturated.
Inhibitors/Enzyme inhibition:
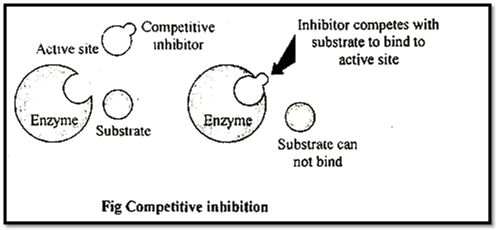
(i) Competitive inhibitors or competitive inhibition and reversible type:
These are substrate analogues, which bind to the active site of enzymes & enzymes get inhibited, such inhibition is called as competitive inhibition.
e.g. Succinic dehydrogenase is inhibited by its competitor malonate,
This is reversible inhibition. Malonate is known as substrate analogue of succlnate.
Similarly sulpha drugs are substrate analogue of p-amino benzoic acid (PABA) used in folic acid synthesis in bacterial cells. Hence these drugs are used to kill bacterial cells.
(ii) Non-competitive inhibitors or non competitive inhibition and irreversible type:
In this type of inhibition, inhibitor substance can bind simultaneously to an enzyme, other than it's active site and destroy the sulfhydril (S-H) group of enzyme.
Example: Toxic metals, CO, CN poisoning of cytochrome oxidase.
Such inhibition are irreversible inhibition.
(iii) Non competitive & reversible type: When inhibitor binds at allosteric site reversibly.
When product of biochemical reaction inhibits the enzyme action, it is known as product inhibition or retro inhibition or feed back inhibition.
The product may binds at allosteric site of allosteric enzyme then it is non-competitive reversible allosteric inhibition.
Example: inhibition of hexokinase by glucose 6P. So, product inhibition is always reversible inhibition.
Also Steric Modulation
Jacob & Monad, first discovered Lvthreonine dehydratase, inhibited by its product isoleucine.
In the allosteric modulation, chemical or products fits in allosteric sites & bring a change in shape of active site of enzyme.
Chemicals which bind at allosteric site of allosteric enzymes are known as allosteric modulators.
These modulators may be activators (positive modulators) or inhibitors (negative modulators) of allosteric enzymes.
If allosteric modulator positively change the configuration of active site, then called positive allosteric modulation and. if negatively change then called negative allosteric modulation respectively by +ve modulator (activators) and-ve modulator (inhibitors) .
Ex. Phosphofructokinase inhibited by. ATP, activated by AMP and ADP.
Some times product also bindsat allosteric site and inhibits the enzyme.
Ex. Hexokinase inhibited by glucose - 6P and exhibits feed back inhibition.
This is a type of reversible and non-competitive inhibition found in allosteric enzymes.
All allosteric modulation are not feed-back inhibition.
Km Constant (Miehalies & Menten Constant): "Km constant of an enzyme, is the concentration of substrate at which rate of reaction of that enzyme attains half of its maximum velocity. It is given by Michaelis & Menten. The value of Km should be lower for an enzyme.
Km exhibits catalytic activity of an enzyme.
Ki constant (Enzyme inhibitor complex dissociation constant) indicates the dissociation of enzyme inhibitor complex (EIC) of reversible inhibitors.
Km and Ki constant of an enzyme should be low.
If competitive inhibitor is present, then km ↑ and Vmax - No change.
If Non-competitive (reversible) inhibitor is present, then,
Km - No change, Vmax - Decrease.
Km =1/2Vmax
Competitive inhibiton is overcome by increase in concentration of substrate.
Km value differs from substrate to substrate because different enzymes differ in their affinity towards different substrates. A high Km indicates low affinity while a low Km shows strong affinity. Protease acts on different proteins. So it's Km value differ from protein to protein.
The Michaelis Menten equation description how reaction relatively varies with substrate concentration as given
V= Vmax/Km
Where V0 is the rate of initial reaction: Vmax is the maximum relative or the reaction rate with excess substrate; Km is the Michaelis constant = K2 + K3/K1:[S] is the substrate concentration.
The above reaction shows that the greater the affinity between an enzyme and its substrate, the lower the Km (in units moles per litre) of the enzyme substrate reaction. Stated inversely, 1 /Km, is the measure of affinity of the enzyme for its substrate.
Enzyme-inhibitor dissociation constant (Ki): it is dissociation constant of enzyme - inhibitor complex.
Ki = [E][I]/[EI]
Where, E is enzyme and I is concentration of inhibitor.
High Ki decreases enzyme activity while low Ki increases some, it is applicable to competitive inhibitors.
To read more, Buy study materials of Biomolecules comprising study notes, revision notes, video lectures, previous year solved questions etc. Also browse for more study materials on Biology here.
View courses by askIITians


Design classes One-on-One in your own way with Top IITians/Medical Professionals
Click Here Know More

Complete Self Study Package designed by Industry Leading Experts
Click Here Know More

Live 1-1 coding classes to unleash the Creator in your Child
Click Here Know More